How to convert my snakemake workflow into a Python package combined to Snakecdysis ?
Generating the python package tree
The first step is to structure your Git repository as a Python package. To help you in this process, you can use the snackecdysis command generate_template with the following parameters:
-p GitHub_Repo_Name. In lower case, corresponding to the name of the repo (e.g.: culebrONT for http://…)
-n package_Name. In lowcase case (python nomenclature) (e.g.: culebrONT )
generate_template -p GitHub_Repo_Name -n package_Name
Running this command generates all the files and directories needed to create a python package. This package is then ready to be used by snakecdysis, simplifying the installation and use of your snakemake pipeline.
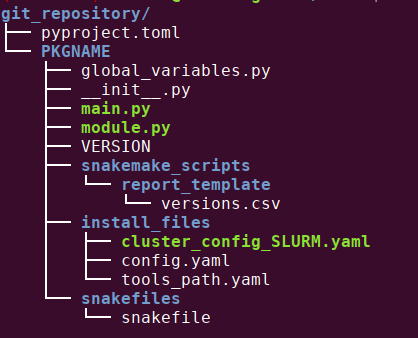
now I will explain what you have to modify in the files to create your package. You must adapt the highlighted lines on example files, to your project.
Configuring your files
Include specific configurations and settings here.
pyproject.toml file
This file is used to create a python package as described here in the official documentation.
click to open pyproject.toml
1# Build system backend to create package to upload on pypi
2[build-system]
3build-backend = "setuptools.build_meta"
4requires = [
5 "setuptools>=68",
6 "setuptools_scm[toml]>=8"
7]
8
9# configuration of setuptools_scm to use tag version autoincrementation
10[tool.setuptools_scm]
11write_to = "PKGNAME/_version.py"
12version_scheme="release-branch-semver"
13tag_regex="^(\\d.\\d.\\d)-*\\w*\\d*$"
14local_scheme = "no-local-version"
15
16# define Project settings
17[project]
18name = "PKGNAME"
19dynamic = ["version"]
20description = "TODO !!!!!"
21authors = [
22 {name = "Ravel Sebastien (CIRAD)",email = "sebastien.ravel@cirad.fr"},
23]
24dependencies = ["PyYAML", "click>=8.0.3", "cookiecutter", "docutils", "python-gitlab", "snakemake<8", "tqdm"]
25requires-python = ">=3.8"
26readme = "README.rst"
27license = {file = "LICENSE"}
28keywords = ["snakemake", "wrapper", "installation"]
29classifiers = [
30 "Development Status :: 5 - Production/Stable",
31 "Intended Audience :: Developers",
32 "Intended Audience :: End Users/Desktop",
33 "License :: CeCILL-C Free Software License Agreement (CECILL-C)",
34 "License :: Free for non-commercial use",
35 "License :: OSI Approved :: MIT License",
36 "Natural Language :: English",
37 "Operating System :: POSIX :: Linux",
38 "Programming Language :: Python :: 3.8",
39 "Programming Language :: Python :: 3.9",
40 "Topic :: Scientific/Engineering",
41 "Topic :: Scientific/Engineering :: Bio-Informatics",
42]
43
44[project.urls]
45Homepage = "https://forge.ird.fr/phim/sravel/snakecdysis"
46Downloads = "https://forge.ird.fr/phim/sravel/snakecdysis/archive/"
47"Bug Tracker" = "https://forge.ird.fr/phim/sravel/snakecdysis/issues"
48Documentation = "https://snakecdysis.readthedocs.io/en/latest/"
49"Source Code" = "https://forge.ird.fr/phim/sravel/snakecdysis"
50
51[project.optional-dependencies]
52dev = [
53 "sphinx_click",
54 "sphinx_copybutton",
55 "sphinx_rtd_theme",
56 "tox",
57]
58
59[project.scripts]
60generate_template = "snakecdysis.scripts.generate_template:main"
61
62[project.entry-points.PKGNAME]
63PKGNAME = "__init__"
64
65[tool.semantic_release]
66logging_use_named_masks = true
67tag_format = "{version}"
68commit_parser = "angular"
69commit_message = "{version}\n\nAutomatically generated by python-semantic-release"
70build_command = """
71 python -m pip install build~=0.10.0
72 python -m build .
73"""
74major_on_zero = true
75assets = []
76version_variables = ["snakecdysis/__init__.py:__version__"]
77version_toml = ["pyproject.toml:project.version"]
78
79[tool.semantic_release.branches.main]
80match = "(main|master)"
81prerelease_token = "rc"
82prerelease = false
83
84[tool.semantic_release.changelog]
85template_dir = "templates"
86changelog_file = "CHANGELOG.md"
87exclude_commit_patterns = []
88
89[tool.semantic_release.changelog.environment]
90block_start_string = "{%"
91block_end_string = "%}"
92variable_start_string = "{{"
93variable_end_string = "}}"
94comment_start_string = "{#"
95comment_end_string = "#}"
96trim_blocks = true
97lstrip_blocks = true
98newline_sequence = "\n"
99keep_trailing_newline = true
100extensions = []
101autoescape = true
102
103[tool.semantic_release.commit_author]
104env = "GIT_COMMIT_AUTHOR"
105default = "semantic-release <semantic-release>"
106
107[tool.semantic_release.commit_parser_options]
108allowed_tags = ["build", "chore", "ci", "docs", "doc", "feat", "fix", "perf", "style", "refactor", "test",
109 "BUILD", "CHORE", "CI", "DOCS", "DOC", "FEAT", "FIX", "PERF", "STYLE", "REFACTOR", "TEST"]
110minor_tags = ["feat", "FEAT"]
111patch_tags = ["fix", "perf", "FIX", "PERF"]
112#
113[tool.semantic_release.remote]
114name = "origin"
115type = "gitlab"
116ignore_token_for_push = false
117token = { env = "GH_TOKEN" }
118domain = "forge.ird.fr"
119api_domain = "forge.ird.fr"
120
121[tool.semantic_release.publish]
122dist_glob_patterns = ["dist/*"]
123upload_to_vcs_release = true
The __init__.py file
This file is the entry point of the python package.
click to open __init__.py
1#!/usr/bin/env python3
2# -*- coding: utf-8 -*-
3from .global_variables import *
4from PKGNAME.module import PKGNAME
5from pathlib import Path
6from .global_variables import GIT_URL, DOCS, DATATEST_URL_FILES, SINGULARITY_URL_FILES
7
8logo = Path(__file__).parent.resolve().joinpath('PKGNAME_logo.png').as_posix()
9
10__version__ = Path(__file__).parent.resolve().joinpath("VERSION").open("r").readline().strip()
11
12
13__doc__ = """BLABLA"""
14
15description_tools = f"""
16 Welcome to PKGNAME version: {__version__} ! Created on XXXX 20XX
17 @author: Sebastien Ravel (CIRAD)
18 @email: Sebastien.ravel@cirad.fr
19
20 Please cite our github: GIT_URL
21 Licencied under MIT and Intellectual property belongs to XXXX and authors.
22 Documentation avail at: DOCS"""
23
24dico_tool = {
25 "soft_path": Path(__file__).resolve().parent.as_posix(),
26 "url": GIT_URL,
27 "docs": DOCS,
28 "description_tool": description_tools,
29 "singularity_url_files": SINGULARITY_URL_FILES,
30 "datatest_url_files": DATATEST_URL_FILES,
31 "snakefile": Path(__file__).resolve().parent.joinpath("snakefiles", "Snakefile"),
32 "snakemake_scripts": Path(__file__).resolve().parent.joinpath("snakemake_scripts")
33}
global_variables.py file
This file allows to group the variables to be used in the wrapper
click to open global_variables.py
1from pathlib import Path
2
3DOCS = "https://PKGNAME.readthedocs.io/en/latest/"
4GIT_URL = "https://github.com/SouthGreenPlatform/PKGNAME"
5
6SINGULARITY_URL_FILES = [('oras://registry.forge.ird.fr/diade/culebront_pipeline/apptainer/apptainer.culebront_tools.sif:0.0.1',
7 f'INSTALL_PATH/containers/apptainer.culebront_tools.sif')
8 ]
9
10DATATEST_URL_FILES = ("https://itrop.ird.fr/culebront_utilities/Data-Xoo-sub.zip", "Data-Xoo-sub.zip")
11
12
13
main.py file
This is the main script of the workflow. You have to check that its path is in the setup.py file. Normally it is already included with the instruction line 75:
entry_points={
'console_scripts': [f"{NAME} = {NAME}.main:main"],
},
This is the main script of the workflow. You have to check that its path is in the setup.py file. Normally it is already included with the instruction line 75:
click to open main.py
1#!/usr/bin/env python3
2
3from snakecdysis import main_wrapper
4from PKGNAME import dico_tool
5
6main = main_wrapper(**dico_tool)
7
8if __name__ == '__main__':
9 main()
Transferring your snakemake workflow
Include blabla on how to transfer your Snakemake workflow into the newly generated Python package.
module.py file
This file is used on snakefile file to add more control of the configuration file and checking user values. The goal is to create a new class that inherits from SnakEcdysis in order to use the attributes in order to have access to, for example, the paths of the scripts, the default/user configuration files, …
click to open module.py
1#!/usr/bin/env python3
2# -*- coding: utf-8 -*-
3from pathlib import Path
4
5from snakemake.logging import logger
6from snakemake.utils import validate
7import re
8from .global_variables import *
9from snakecdysis import *
10
11
12class PKGNAME(SnakEcdysis):
13 """
14 to read file config
15 """
16
17 def __init__(self, dico_tool, workflow, config):
18 super().__init__(**dico_tool, workflow=workflow, config=config)
19 # workflow is available only in __init__
20 # print("\n".join(list(workflow.__dict__.keys())))
21 # print(workflow.__dict__)
22
23 # Initialisation of PKGNAME attributes
snakefile file
click to open snakefile
1#!/usr/bin/env snakemake
2# -*- coding: utf-8 -*-
3
4from pathlib import Path
5from pprint import pprint as pp
6# load own functions
7import PKGNAME
8
9
10PKGNAME_obj = PKGNAME(PKGNAME.dico_tool, workflow=workflow, config=config)
11tools_config = PKGNAME_obj.tools_config
12cluster_config = PKGNAME_obj.cluster_config
13
14# print(PKGNAME_obj.export_use_yaml)
15# print for debug:
16# pp(PKGNAME_obj)
17# exit()
18# print(tools_config)
19# exit()
20
21###############################################################################
22# dir and suffix
23output_dir = config["DATA"]["OUTPUT"]
24log_dir = f"{output_dir}LOGS/"
25
26# Change workdir to output path (slurm logs append on outdir)
27workdir: output_dir
28
29
30
31#############################################
32# use threads define in cluster_config rule or rule default or default in snakefile
33#############################################
34def get_threads(rule, default):
35 """
36 give threads or 'cpus-per-task from cluster_config rule: threads to SGE and cpus-per-task to SLURM
37 """
38 if cluster_config:
39 if rule in cluster_config and 'threads' in cluster_config[rule]:
40 return int(cluster_config[rule]['threads'])
41 elif rule in cluster_config and 'cpus-per-task' in cluster_config[rule]:
42 return int(cluster_config[rule]['cpus-per-task'])
43 elif '__default__' in cluster_config and 'cpus-per-task' in cluster_config['__default__']:
44 return int(cluster_config['__default__']['cpus-per-task'])
45 elif '__default__' in cluster_config and 'threads' in cluster_config['__default__']:
46 return int(cluster_config['__default__']['threads'])
47 # if local
48 elif workflow.global_resources["_cores"]:
49 return workflow.global_resources["_cores"]
50 # if cluster not rule and not default or local not _cores return value from call
51 return default
52
53
54
55################################ ASSEMBLY ####################################
56include: f"{PKGNAME_obj.install_path}/snakefiles/assemblers.snake"
57
58
59
60rule some_exemple:
61 threads: get_threads("some_exemple",1)
62 input:
63 toto=rules.start.output.machin,
64 truc=ref
65 output:
66 csv_var_per_contig=f"{output_dir}4_STRUCTURAL_VAR/csv_variants/{{samples}}_variants_per_contig.csv"
67 log:
68 error=f'{log_dir}some_exemple/{{samples}}.e',
69 output=f'{log_dir}some_exemple/{{samples}}.o'
70 message:
71 f"""
72 Running {{rule}}
73 Input:
74 - toto: {{input.toto}}
75 Output:
76 - csv_file: {{output.csv_var_per_contig}}
77 Others
78 - Threads: {{threads}}
79 - LOG error: {{log.error}}
80 - LOG output: {{log.output}}
81 """
82 singularity:
83 tools_config['SINGULARITY']['TOOLS']
84 envmodules:
85 tools_config["MODULES"]["MINIMAP2"],
86 tools_config["MODULES"]["SAMTOOLS"]
87 shell:
88 f"(python {PKGNAME_obj.snakemake_scripts}/vcf_contigs.py -v {{input.vcf_file}} -r {{input.ref_fasta}} -o {{output.csv_var_per_contig}}) 1>{{log.output}} 2>{{log.error}}"
89
90
91
update documentation comming soon
# USER manual - How to install my workflow after developping the python package of my workflow